Exploratory Data Analysis (EDA) Wine Quality dataset
Contents
Exploratory Data Analysis (EDA) Wine Quality dataset#
We will analyze the well-known wine dataset using our newly gained skills in this part. Each wine is described with several attributes obtained by physicochemical tests and by its quality (from 1 to 10). This dataset is perfect for many ML tasks such as:
Testing Outlier detection algorithms that can detect the few excellent or poor wines
Modeling the relationship between wine quality and wine attributes (using regression or classification)
Attribute (feature) selection techniques to remove unimportant features
✏️ The example is inspired by [Pac].
Let’s start by importing packages:
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
Let’s load our data (there are two datasets for red and white wines):
red_wine_url = "https://archive.ics.uci.edu/ml/machine-learning-databases/wine-quality/winequality-red.csv"
white_wine_url = "https://archive.ics.uci.edu/ml/machine-learning-databases/wine-quality/winequality-white.csv"
# load data to DF
df_red = pd.read_csv(red_wine_url, delimiter=";")
df_white = pd.read_csv(white_wine_url, delimiter=";")
Let’s check red wine data:
df_red.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 1599 entries, 0 to 1598
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 fixed acidity 1599 non-null float64
1 volatile acidity 1599 non-null float64
2 citric acid 1599 non-null float64
3 residual sugar 1599 non-null float64
4 chlorides 1599 non-null float64
5 free sulfur dioxide 1599 non-null float64
6 total sulfur dioxide 1599 non-null float64
7 density 1599 non-null float64
8 pH 1599 non-null float64
9 sulphates 1599 non-null float64
10 alcohol 1599 non-null float64
11 quality 1599 non-null int64
dtypes: float64(11), int64(1)
memory usage: 150.0 KB
df_red.describe()
| fixed acidity | volatile acidity | citric acid | residual sugar | chlorides | free sulfur dioxide | total sulfur dioxide | density | pH | sulphates | alcohol | quality | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 1599.000000 | 1599.000000 | 1599.000000 | 1599.000000 | 1599.000000 | 1599.000000 | 1599.000000 | 1599.000000 | 1599.000000 | 1599.000000 | 1599.000000 | 1599.000000 |
| mean | 8.319637 | 0.527821 | 0.270976 | 2.538806 | 0.087467 | 15.874922 | 46.467792 | 0.996747 | 3.311113 | 0.658149 | 10.422983 | 5.636023 |
| std | 1.741096 | 0.179060 | 0.194801 | 1.409928 | 0.047065 | 10.460157 | 32.895324 | 0.001887 | 0.154386 | 0.169507 | 1.065668 | 0.807569 |
| min | 4.600000 | 0.120000 | 0.000000 | 0.900000 | 0.012000 | 1.000000 | 6.000000 | 0.990070 | 2.740000 | 0.330000 | 8.400000 | 3.000000 |
| 25% | 7.100000 | 0.390000 | 0.090000 | 1.900000 | 0.070000 | 7.000000 | 22.000000 | 0.995600 | 3.210000 | 0.550000 | 9.500000 | 5.000000 |
| 50% | 7.900000 | 0.520000 | 0.260000 | 2.200000 | 0.079000 | 14.000000 | 38.000000 | 0.996750 | 3.310000 | 0.620000 | 10.200000 | 6.000000 |
| 75% | 9.200000 | 0.640000 | 0.420000 | 2.600000 | 0.090000 | 21.000000 | 62.000000 | 0.997835 | 3.400000 | 0.730000 | 11.100000 | 6.000000 |
| max | 15.900000 | 1.580000 | 1.000000 | 15.500000 | 0.611000 | 72.000000 | 289.000000 | 1.003690 | 4.010000 | 2.000000 | 14.900000 | 8.000000 |
Let’s check white wine data:
df_white.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 4898 entries, 0 to 4897
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 fixed acidity 4898 non-null float64
1 volatile acidity 4898 non-null float64
2 citric acid 4898 non-null float64
3 residual sugar 4898 non-null float64
4 chlorides 4898 non-null float64
5 free sulfur dioxide 4898 non-null float64
6 total sulfur dioxide 4898 non-null float64
7 density 4898 non-null float64
8 pH 4898 non-null float64
9 sulphates 4898 non-null float64
10 alcohol 4898 non-null float64
11 quality 4898 non-null int64
dtypes: float64(11), int64(1)
memory usage: 459.3 KB
df_white.describe()
| fixed acidity | volatile acidity | citric acid | residual sugar | chlorides | free sulfur dioxide | total sulfur dioxide | density | pH | sulphates | alcohol | quality | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 4898.000000 | 4898.000000 | 4898.000000 | 4898.000000 | 4898.000000 | 4898.000000 | 4898.000000 | 4898.000000 | 4898.000000 | 4898.000000 | 4898.000000 | 4898.000000 |
| mean | 6.854788 | 0.278241 | 0.334192 | 6.391415 | 0.045772 | 35.308085 | 138.360657 | 0.994027 | 3.188267 | 0.489847 | 10.514267 | 5.877909 |
| std | 0.843868 | 0.100795 | 0.121020 | 5.072058 | 0.021848 | 17.007137 | 42.498065 | 0.002991 | 0.151001 | 0.114126 | 1.230621 | 0.885639 |
| min | 3.800000 | 0.080000 | 0.000000 | 0.600000 | 0.009000 | 2.000000 | 9.000000 | 0.987110 | 2.720000 | 0.220000 | 8.000000 | 3.000000 |
| 25% | 6.300000 | 0.210000 | 0.270000 | 1.700000 | 0.036000 | 23.000000 | 108.000000 | 0.991723 | 3.090000 | 0.410000 | 9.500000 | 5.000000 |
| 50% | 6.800000 | 0.260000 | 0.320000 | 5.200000 | 0.043000 | 34.000000 | 134.000000 | 0.993740 | 3.180000 | 0.470000 | 10.400000 | 6.000000 |
| 75% | 7.300000 | 0.320000 | 0.390000 | 9.900000 | 0.050000 | 46.000000 | 167.000000 | 0.996100 | 3.280000 | 0.550000 | 11.400000 | 6.000000 |
| max | 14.200000 | 1.100000 | 1.660000 | 65.800000 | 0.346000 | 289.000000 | 440.000000 | 1.038980 | 3.820000 | 1.080000 | 14.200000 | 9.000000 |
As we will later combine our datasets into a single one, let’s add an extra column to indicate category:
df_white['wine_category'] = 'white'
df_red['wine_category'] = 'red'
A colleague in our department, a wine expert, gave us a tip that scaling the quality from 1 to 10 might probably be too granular for certain types of analysis, and it might be OK to divide wines into three quality categories - low, medium, high:
df_red['quality_label'] = df_red['quality'].apply(lambda value: ('low' if value <= 5 else 'medium') if value <= 7 else 'high')
df_red['quality_label'] = pd.Categorical(df_red['quality_label'], categories=['low', 'medium', 'high'])
df_white['quality_label'] = df_white['quality'].apply(lambda value: ('low' if value <= 5 else 'medium') if value <= 7 else 'high')
df_white['quality_label'] = pd.Categorical(df_white['quality_label'], categories=['low', 'medium', 'high'])
Red Wine Dataset Analysis#
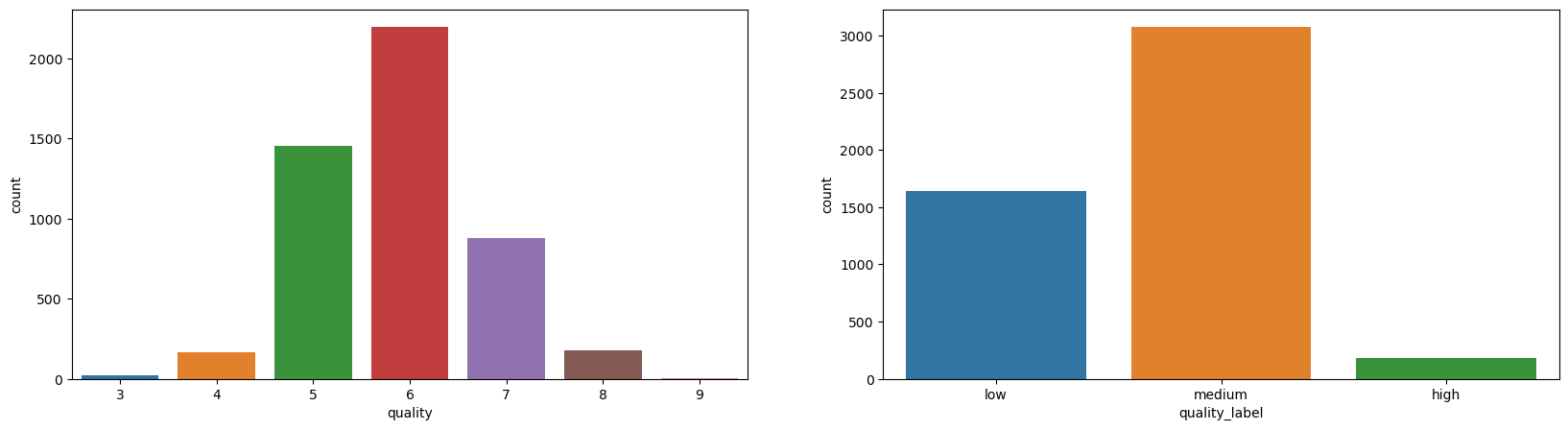
Let’s get familiar with the red wine dataset with some visualizations that we already know:
_, axes = plt.subplots(1, 2, figsize=(20, 5))
sns.countplot(data=df_red, x='quality', ax=axes[0])
sns.countplot(data=df_red, x='quality_label', ax=axes[1]);
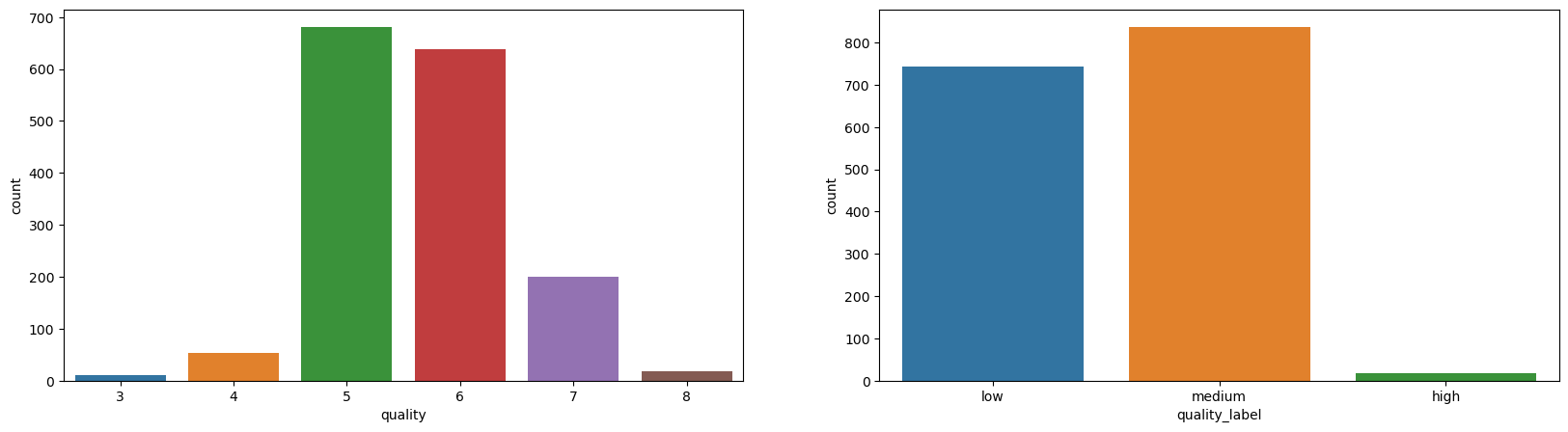
Well, the dataset is highly imbalanced when it comes to quality attribute.
Let’s check some relationships between attributes:
sns.pairplot(df_red.drop(columns=['quality']), hue="quality_label", height=3);
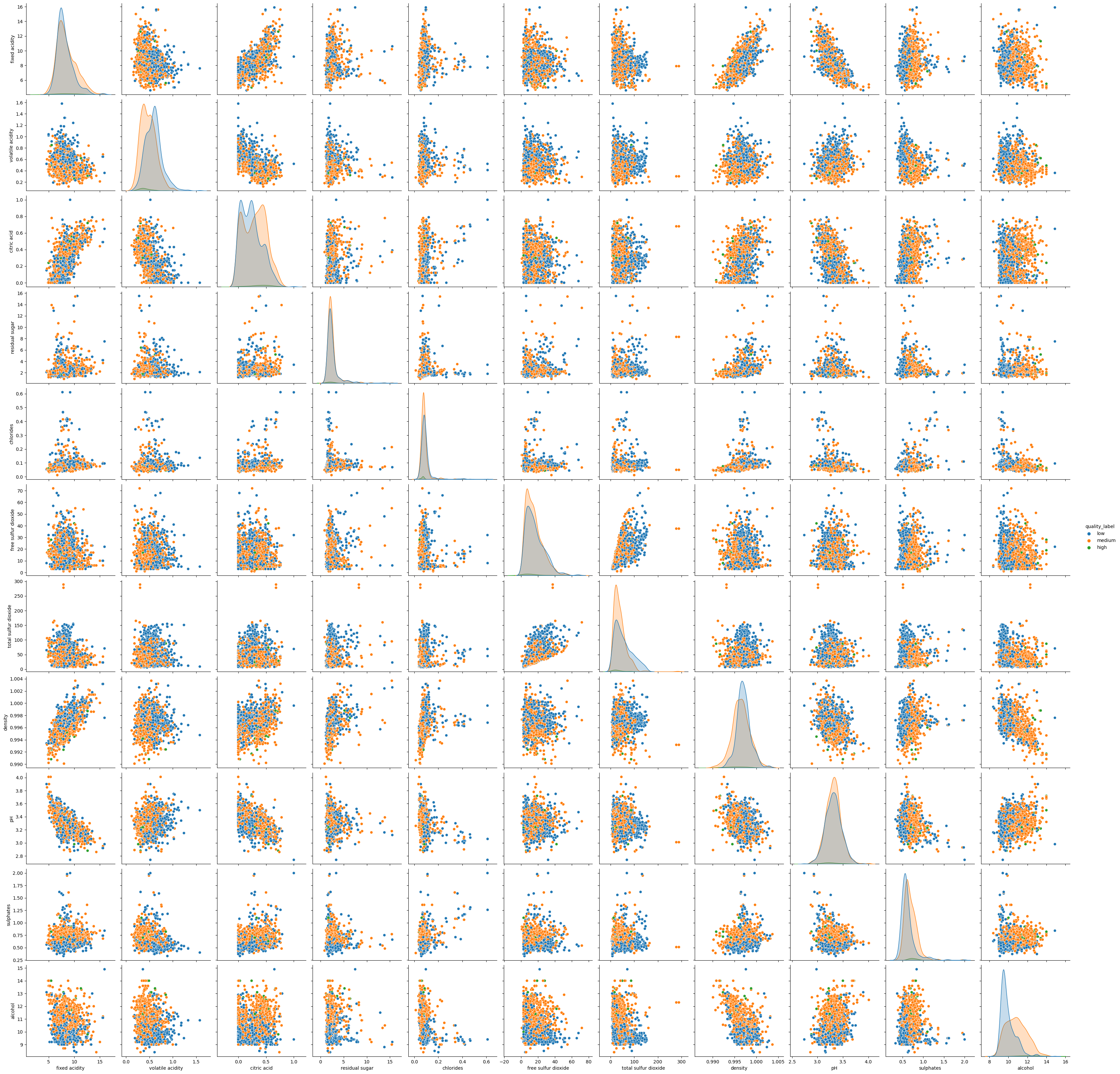
The plot is somehow hard to read. Let’s try to focus on an individual attribute:
_, axes = plt.subplots(1, 2, figsize=(10, 5))
sns.violinplot(data=df_red, y="alcohol", x="quality_label", ax=axes[0])
sns.violinplot(data=df_red, y="alcohol", x="quality", ax=axes[1]);
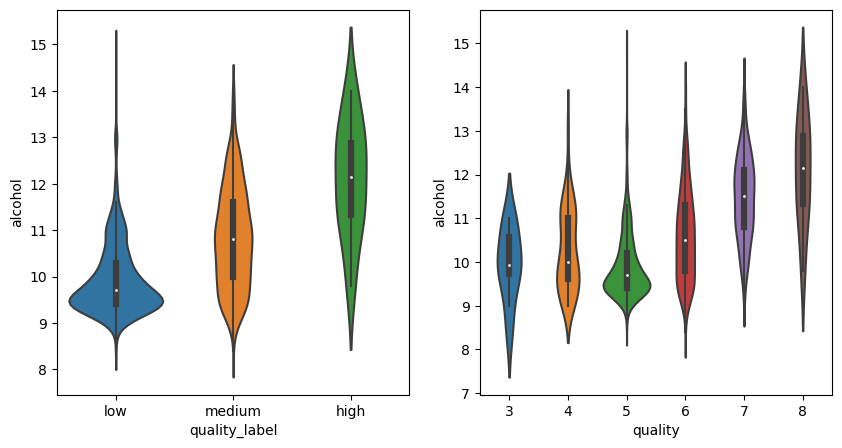
There are no obvious gotchas. Maybe we will be more lucky with the correlation plot:
sns.heatmap(df_red.corr(method="pearson"), annot=True);
/tmp/ipykernel_18914/527241991.py:1: FutureWarning: The default value of numeric_only in DataFrame.corr is deprecated. In a future version, it will default to False. Select only valid columns or specify the value of numeric_only to silence this warning.
sns.heatmap(df_red.corr(method="pearson"), annot=True);
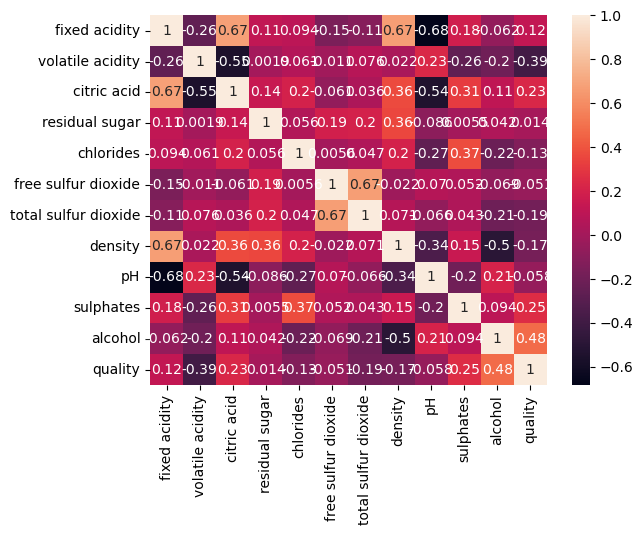
There are definitely some interactions, we can study them, for example, with jointplot:
sns.jointplot(x='alcohol',y='sulphates',data=df_red, kind='reg');
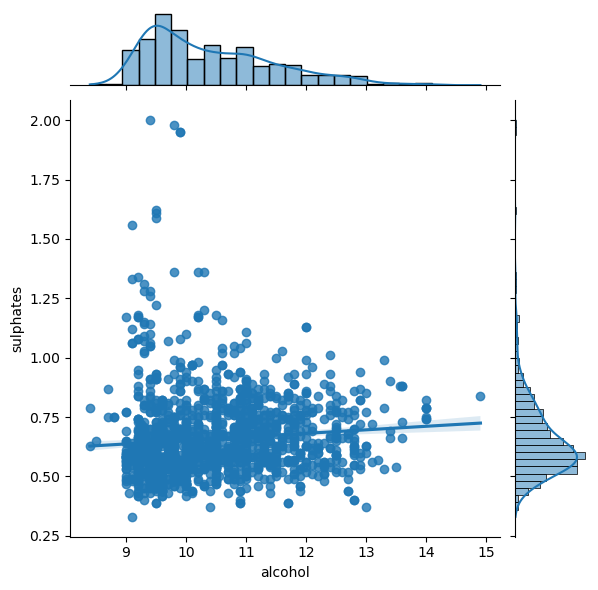
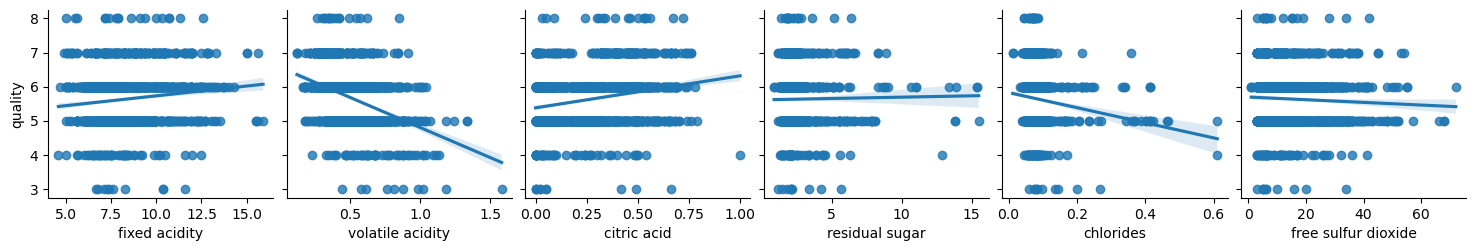
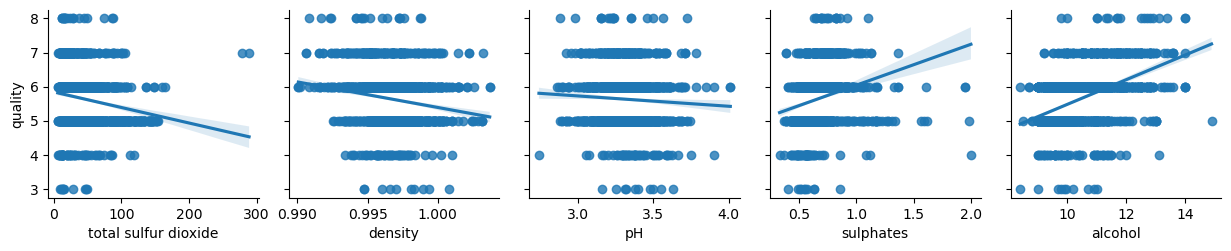
Let’s analyze relationship between attributes and the quality:
columns = list(filter(lambda col: col not in {'quality', 'wine_category', 'quality_label'}, df_red.columns))
chunks = [columns[i:i + 6] for i in range(0, len(columns), 6)]
for columns_names in chunks:
sns.pairplot(df_red, x_vars=columns_names, y_vars='quality', kind="reg")
White Wine Dataset Analysis#
Let’s run the same analysis for the white vine:
_, axes = plt.subplots(1, 2, figsize=(20, 5))
sns.countplot(data=df_white, x='quality', ax=axes[0])
sns.countplot(data=df_white, x='quality_label', ax=axes[1]);
sns.pairplot(df_white.drop(columns=['quality']), hue="quality_label", height=3);
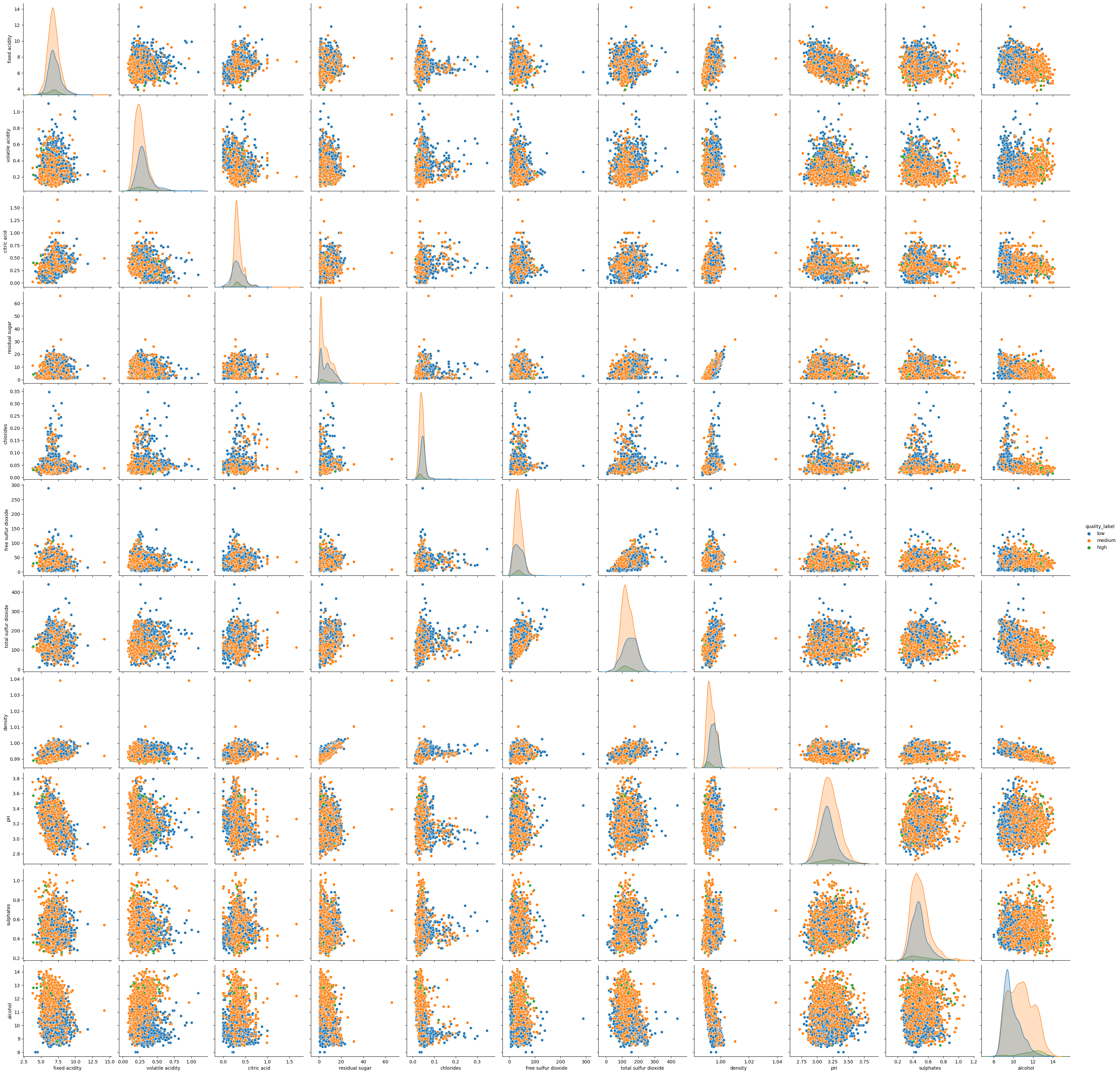
_, axes = plt.subplots(1, 2, figsize=(10, 5))
sns.violinplot(data=df_white, y="alcohol", x="quality_label", ax=axes[0])
sns.violinplot(data=df_white, y="alcohol", x="quality", ax=axes[1]);
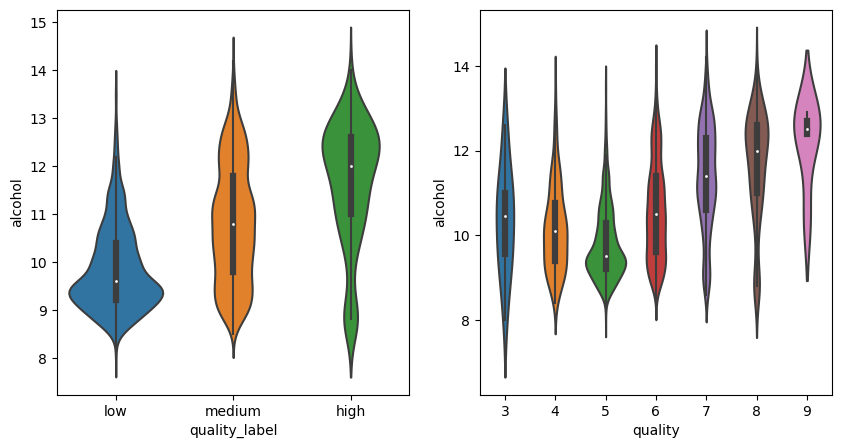
sns.heatmap(df_white.corr(method="pearson"), annot=True);
/tmp/ipykernel_18914/455693947.py:1: FutureWarning: The default value of numeric_only in DataFrame.corr is deprecated. In a future version, it will default to False. Select only valid columns or specify the value of numeric_only to silence this warning.
sns.heatmap(df_white.corr(method="pearson"), annot=True);
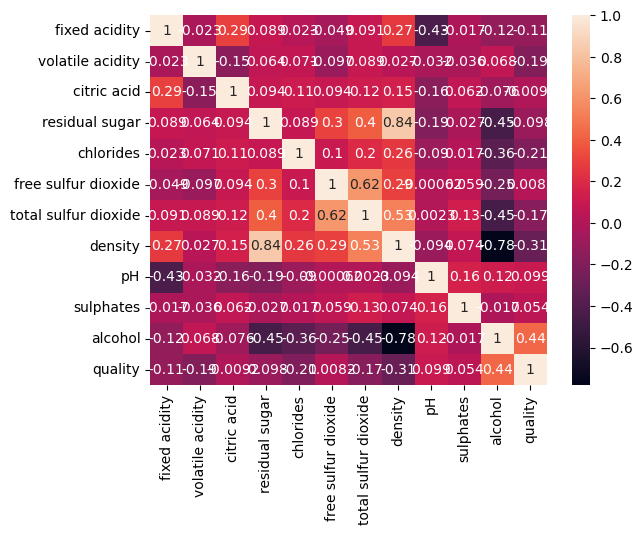
sns.jointplot(x='alcohol',y='sulphates',data=df_white, kind='reg');
columns = list(filter(lambda col: col not in {'quality', 'wine_category', 'quality_label'}, df_white.columns))
chunks = [columns[i:i + 6] for i in range(0, len(columns), 6)]
for columns_names in chunks:
sns.pairplot(df_white, x_vars=columns_names, y_vars='quality', kind="reg")
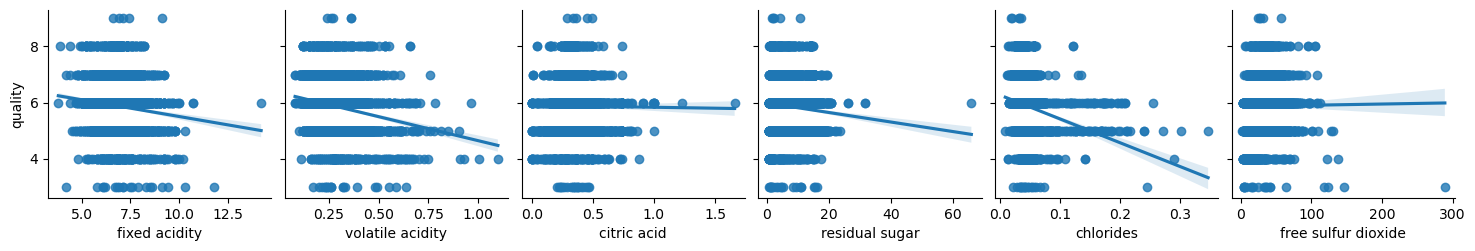
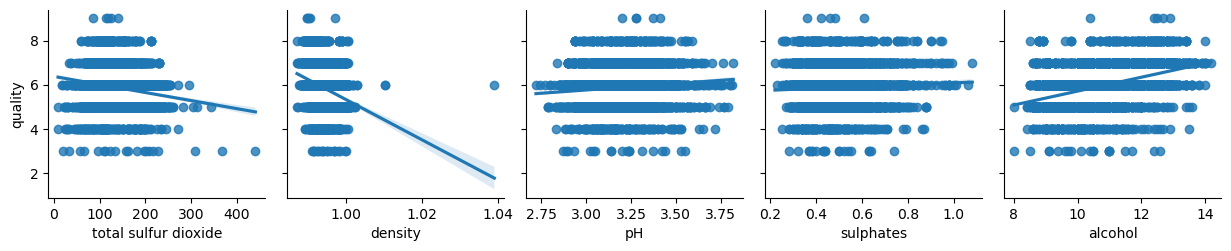
Not surprisingly enough, white wine has a bit of different properties. Let’s combine datasets for additional analysis.
Analyze Combined Data#
Let’s concatenate datasets together:
df_wines = pd.concat([df_red, df_white])
# re-shuffle records just to randomize data points
df_wines = df_wines.sample(frac=1.0, random_state=42).reset_index(drop=True)
df_wines.head(10)
| fixed acidity | volatile acidity | citric acid | residual sugar | chlorides | free sulfur dioxide | total sulfur dioxide | density | pH | sulphates | alcohol | quality | wine_category | quality_label | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 7.0 | 0.17 | 0.74 | 12.8 | 0.045 | 24.0 | 126.0 | 0.99420 | 3.26 | 0.38 | 12.2 | 8 | white | high |
| 1 | 7.7 | 0.64 | 0.21 | 2.2 | 0.077 | 32.0 | 133.0 | 0.99560 | 3.27 | 0.45 | 9.9 | 5 | red | low |
| 2 | 6.8 | 0.39 | 0.34 | 7.4 | 0.020 | 38.0 | 133.0 | 0.99212 | 3.18 | 0.44 | 12.0 | 7 | white | medium |
| 3 | 6.3 | 0.28 | 0.47 | 11.2 | 0.040 | 61.0 | 183.0 | 0.99592 | 3.12 | 0.51 | 9.5 | 6 | white | medium |
| 4 | 7.4 | 0.35 | 0.20 | 13.9 | 0.054 | 63.0 | 229.0 | 0.99888 | 3.11 | 0.50 | 8.9 | 6 | white | medium |
| 5 | 7.2 | 0.53 | 0.14 | 2.1 | 0.064 | 15.0 | 29.0 | 0.99323 | 3.35 | 0.61 | 12.1 | 6 | red | medium |
| 6 | 7.5 | 0.27 | 0.31 | 17.7 | 0.051 | 33.0 | 173.0 | 0.99900 | 3.09 | 0.64 | 10.2 | 5 | white | low |
| 7 | 6.8 | 0.11 | 0.27 | 8.6 | 0.044 | 45.0 | 104.0 | 0.99454 | 3.20 | 0.37 | 9.9 | 6 | white | medium |
| 8 | 9.0 | 0.44 | 0.49 | 2.4 | 0.078 | 26.0 | 121.0 | 0.99780 | 3.23 | 0.58 | 9.2 | 5 | red | low |
| 9 | 7.1 | 0.23 | 0.30 | 2.6 | 0.034 | 62.0 | 148.0 | 0.99121 | 3.03 | 0.56 | 11.3 | 7 | white | medium |
Just by comparing the quality, you can notice a different distributions. For example, the proportion of 5 and 6 is very different for red and white:
_, axes = plt.subplots(1, 2, figsize=(20, 5))
sns.countplot(data=df_wines, x="quality", hue="wine_category", ax=axes[0])
sns.countplot(data=df_wines, x="quality_label", hue="wine_category", ax=axes[1]);
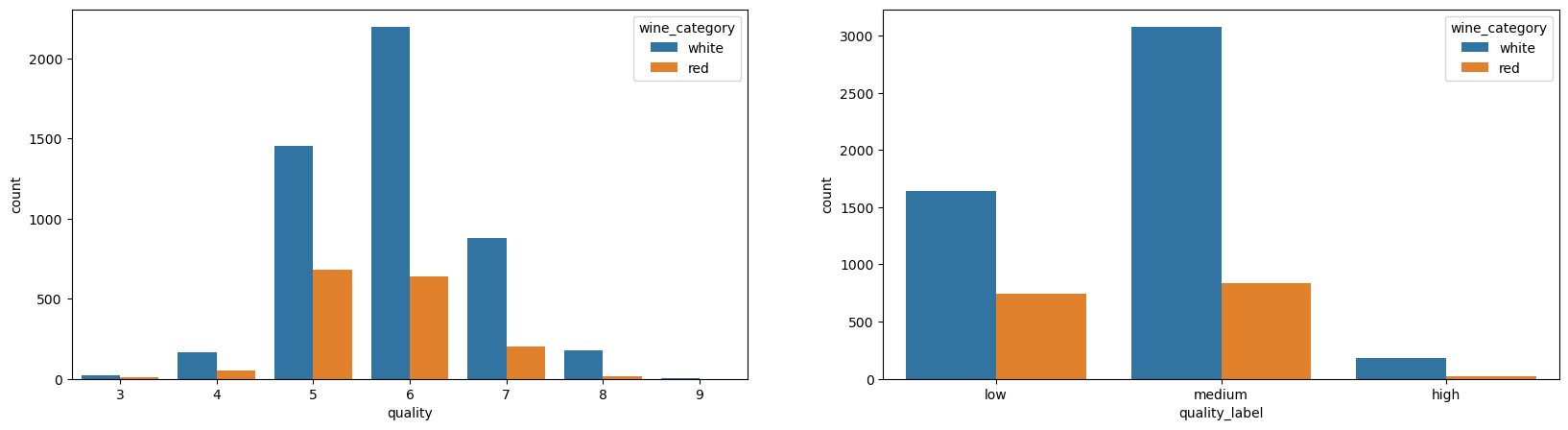
sns.pairplot(df_wines.drop(columns=['quality']), hue="wine_category", height=3);
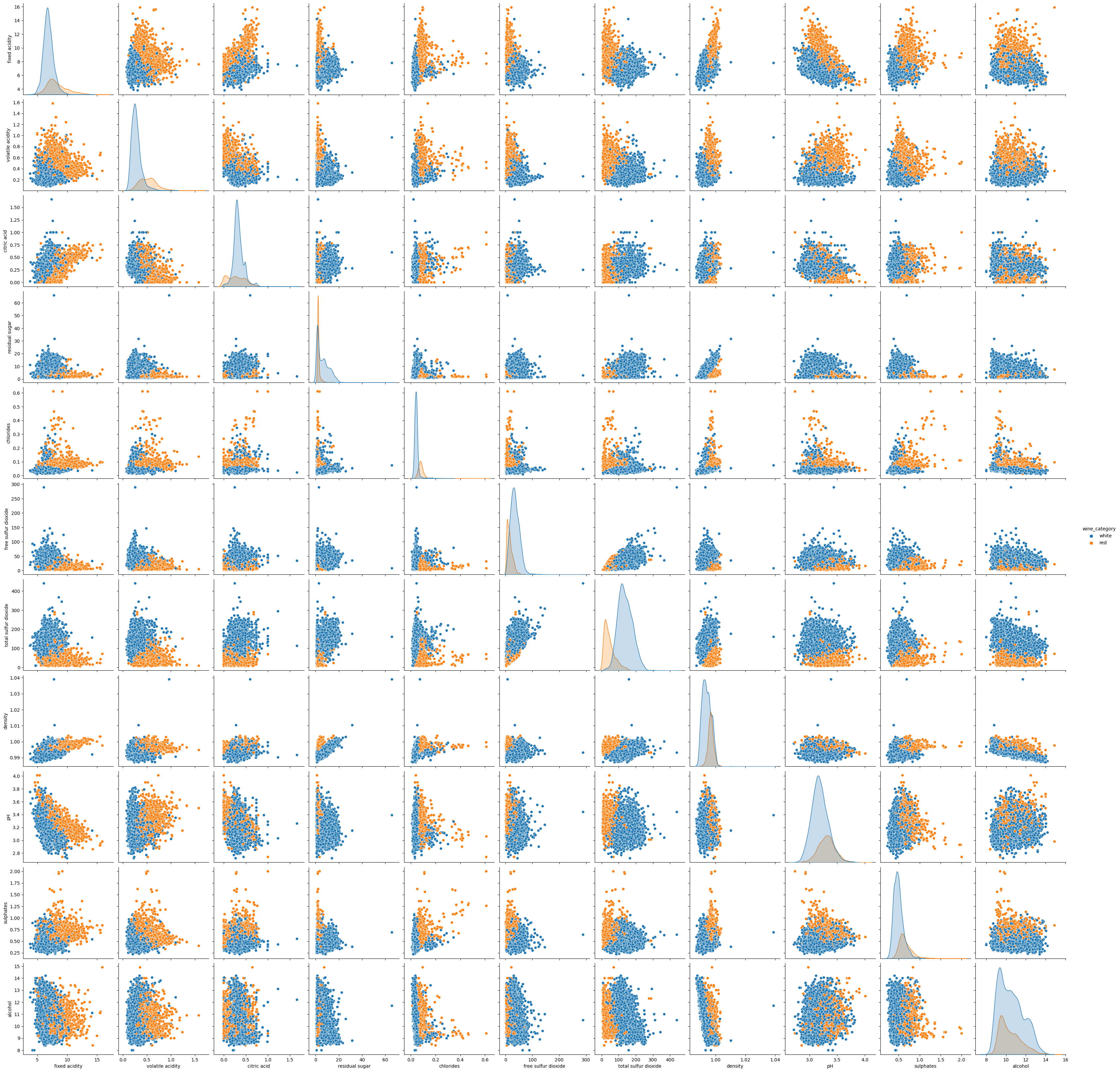
It seems that separating type of wines might be an easy task to do. Let’s check some interactions of combined dataset vs. quality:
sns.pairplot(df_wines.drop(columns=['quality']), hue="quality_label", height=3);
Error in callback <function flush_figures at 0x7f6dc92dd160> (for post_execute):
---------------------------------------------------------------------------
KeyboardInterrupt Traceback (most recent call last)
File ~/miniconda3/envs/ds-academy-development/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py:126, in flush_figures()
123 if InlineBackend.instance().close_figures:
124 # ignore the tracking, just draw and close all figures
125 try:
--> 126 return show(True)
127 except Exception as e:
128 # safely show traceback if in IPython, else raise
129 ip = get_ipython()
File ~/miniconda3/envs/ds-academy-development/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py:90, in show(close, block)
88 try:
89 for figure_manager in Gcf.get_all_fig_managers():
---> 90 display(
91 figure_manager.canvas.figure,
92 metadata=_fetch_figure_metadata(figure_manager.canvas.figure)
93 )
94 finally:
95 show._to_draw = []
File ~/miniconda3/envs/ds-academy-development/lib/python3.9/site-packages/IPython/core/display_functions.py:298, in display(include, exclude, metadata, transient, display_id, raw, clear, *objs, **kwargs)
296 publish_display_data(data=obj, metadata=metadata, **kwargs)
297 else:
--> 298 format_dict, md_dict = format(obj, include=include, exclude=exclude)
299 if not format_dict:
300 # nothing to display (e.g. _ipython_display_ took over)
301 continue
File ~/miniconda3/envs/ds-academy-development/lib/python3.9/site-packages/IPython/core/formatters.py:178, in DisplayFormatter.format(self, obj, include, exclude)
176 md = None
177 try:
--> 178 data = formatter(obj)
179 except:
180 # FIXME: log the exception
181 raise
File ~/miniconda3/envs/ds-academy-development/lib/python3.9/site-packages/decorator.py:232, in decorate.<locals>.fun(*args, **kw)
230 if not kwsyntax:
231 args, kw = fix(args, kw, sig)
--> 232 return caller(func, *(extras + args), **kw)
File ~/miniconda3/envs/ds-academy-development/lib/python3.9/site-packages/IPython/core/formatters.py:222, in catch_format_error(method, self, *args, **kwargs)
220 """show traceback on failed format call"""
221 try:
--> 222 r = method(self, *args, **kwargs)
223 except NotImplementedError:
224 # don't warn on NotImplementedErrors
225 return self._check_return(None, args[0])
File ~/miniconda3/envs/ds-academy-development/lib/python3.9/site-packages/IPython/core/formatters.py:339, in BaseFormatter.__call__(self, obj)
337 pass
338 else:
--> 339 return printer(obj)
340 # Finally look for special method names
341 method = get_real_method(obj, self.print_method)
File ~/miniconda3/envs/ds-academy-development/lib/python3.9/site-packages/IPython/core/pylabtools.py:151, in print_figure(fig, fmt, bbox_inches, base64, **kwargs)
148 from matplotlib.backend_bases import FigureCanvasBase
149 FigureCanvasBase(fig)
--> 151 fig.canvas.print_figure(bytes_io, **kw)
152 data = bytes_io.getvalue()
153 if fmt == 'svg':
File ~/miniconda3/envs/ds-academy-development/lib/python3.9/site-packages/matplotlib/backend_bases.py:2338, in FigureCanvasBase.print_figure(self, filename, dpi, facecolor, edgecolor, orientation, format, bbox_inches, pad_inches, bbox_extra_artists, backend, **kwargs)
2334 try:
2335 # _get_renderer may change the figure dpi (as vector formats
2336 # force the figure dpi to 72), so we need to set it again here.
2337 with cbook._setattr_cm(self.figure, dpi=dpi):
-> 2338 result = print_method(
2339 filename,
2340 facecolor=facecolor,
2341 edgecolor=edgecolor,
2342 orientation=orientation,
2343 bbox_inches_restore=_bbox_inches_restore,
2344 **kwargs)
2345 finally:
2346 if bbox_inches and restore_bbox:
File ~/miniconda3/envs/ds-academy-development/lib/python3.9/site-packages/matplotlib/backend_bases.py:2204, in FigureCanvasBase._switch_canvas_and_return_print_method.<locals>.<lambda>(*args, **kwargs)
2200 optional_kws = { # Passed by print_figure for other renderers.
2201 "dpi", "facecolor", "edgecolor", "orientation",
2202 "bbox_inches_restore"}
2203 skip = optional_kws - {*inspect.signature(meth).parameters}
-> 2204 print_method = functools.wraps(meth)(lambda *args, **kwargs: meth(
2205 *args, **{k: v for k, v in kwargs.items() if k not in skip}))
2206 else: # Let third-parties do as they see fit.
2207 print_method = meth
File ~/miniconda3/envs/ds-academy-development/lib/python3.9/site-packages/matplotlib/_api/deprecation.py:410, in delete_parameter.<locals>.wrapper(*inner_args, **inner_kwargs)
400 deprecation_addendum = (
401 f"If any parameter follows {name!r}, they should be passed as "
402 f"keyword, not positionally.")
403 warn_deprecated(
404 since,
405 name=repr(name),
(...)
408 else deprecation_addendum,
409 **kwargs)
--> 410 return func(*inner_args, **inner_kwargs)
File ~/miniconda3/envs/ds-academy-development/lib/python3.9/site-packages/matplotlib/backends/backend_agg.py:520, in FigureCanvasAgg.print_png(self, filename_or_obj, metadata, pil_kwargs, *args)
471 @_api.delete_parameter("3.5", "args")
472 def print_png(self, filename_or_obj, *args,
473 metadata=None, pil_kwargs=None):
474 """
475 Write the figure to a PNG file.
476
(...)
518 *metadata*, including the default 'Software' key.
519 """
--> 520 self._print_pil(filename_or_obj, "png", pil_kwargs, metadata)
File ~/miniconda3/envs/ds-academy-development/lib/python3.9/site-packages/matplotlib/backends/backend_agg.py:467, in FigureCanvasAgg._print_pil(self, filename_or_obj, fmt, pil_kwargs, metadata)
462 """
463 Draw the canvas, then save it using `.image.imsave` (to which
464 *pil_kwargs* and *metadata* are forwarded).
465 """
466 FigureCanvasAgg.draw(self)
--> 467 mpl.image.imsave(
468 filename_or_obj, self.buffer_rgba(), format=fmt, origin="upper",
469 dpi=self.figure.dpi, metadata=metadata, pil_kwargs=pil_kwargs)
File ~/miniconda3/envs/ds-academy-development/lib/python3.9/site-packages/matplotlib/image.py:1656, in imsave(fname, arr, vmin, vmax, cmap, format, origin, dpi, metadata, pil_kwargs)
1654 pil_kwargs.setdefault("format", format)
1655 pil_kwargs.setdefault("dpi", (dpi, dpi))
-> 1656 image.save(fname, **pil_kwargs)
File ~/miniconda3/envs/ds-academy-development/lib/python3.9/site-packages/PIL/Image.py:2284, in Image.save(self, fp, format, **params)
2281 filename = fp.name
2283 # may mutate self!
-> 2284 self._ensure_mutable()
2286 save_all = params.pop("save_all", False)
2287 self.encoderinfo = params
File ~/miniconda3/envs/ds-academy-development/lib/python3.9/site-packages/PIL/Image.py:599, in Image._ensure_mutable(self)
597 def _ensure_mutable(self):
598 if self.readonly:
--> 599 self._copy()
600 else:
601 self.load()
File ~/miniconda3/envs/ds-academy-development/lib/python3.9/site-packages/PIL/Image.py:593, in Image._copy(self)
591 def _copy(self):
592 self.load()
--> 593 self.im = self.im.copy()
594 self.pyaccess = None
595 self.readonly = 0
KeyboardInterrupt:
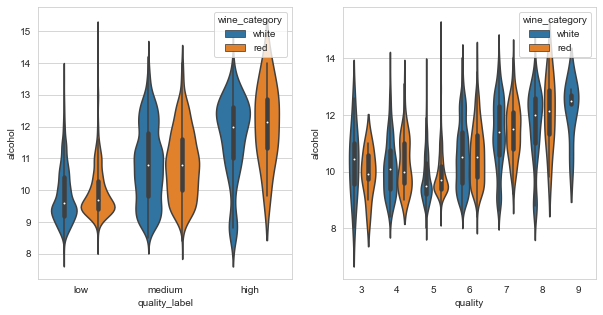
_, axes = plt.subplots(1, 2, figsize=(10, 5))
sns.violinplot(data=df_wines, y="alcohol", x="quality_label", hue="wine_category", ax=axes[0])
sns.violinplot(data=df_wines, y="alcohol", x="quality", hue="wine_category", ax=axes[1]);
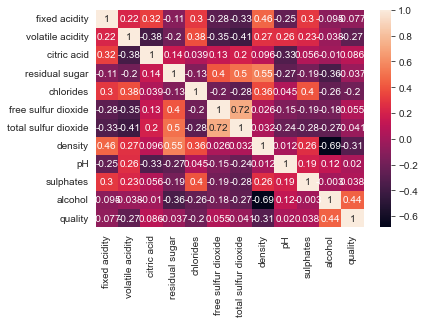
sns.heatmap(df_wines.corr(method="pearson"), annot=True);
columns = list(filter(lambda col: col not in {'quality', 'wine_category', 'quality_label'}, df_wines.columns))
chunks = [columns[i:i + 6] for i in range(0, len(columns), 6)]
for columns_names in chunks:
sns.pairplot(df_wines, x_vars=columns_names, y_vars='quality', kind="reg")
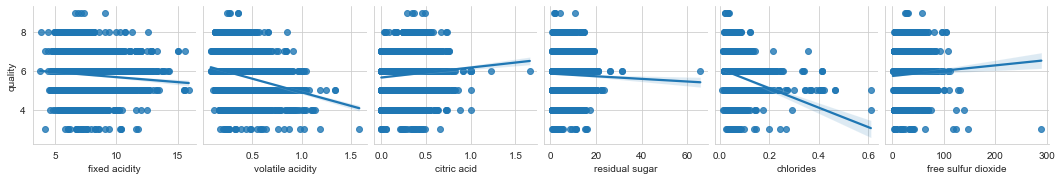
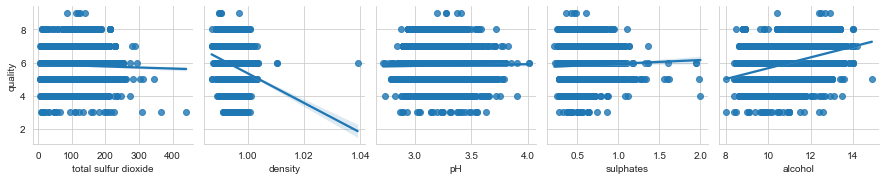
To revisit ML tasks mentioned earlier - it is tough to tell if it is better to solve those tasks using individual datasets or the combined one based on the analysis. My bet would be to do it on individual ones, but it depends on what you want to solve and which approach you take. As always, you need to experiment. I can imagine a scenario where, for example, the smaller white wines dataset can benefit from additional data from the red wines.
✏️ Several libraries aim to automate EDA to some extent if you want to get some results as quickly as possible. One of them is pandas-profiling. An example of automated analysis is Census Dataset EDA.
Exercises#
What sorts of passengers were more likely to survive the Titanic crash?#
For the exercise, you will use the famous Titanic dataset. The sinking of the Titanic is one of the most infamous shipwrecks in history.
Unfortunately, there weren’t enough lifeboats for everyone onboard on Titanic, resulting in 1502 out of 2224 passengers and crew deaths. While some elements of luck were involved in surviving, some groups of people seemed more likely to survive than others.
# load the dataset
titanic_url = 'https://web.stanford.edu/class/archive/cs/cs109/cs109.1166/stuff/titanic.csv'
titanic_data = pd.read_csv(titanic_url)
# TODO: your answer here
Resources#
- Pac
PacktPublishing. Packtpublishing/hands-on-exploratory-data-analysis-with-python: hands-on exploratory data analysis with python, published by packt. URL: https://github.com/PacktPublishing/Hands-on-Exploratory-Data-Analysis-with-Python.

